Our research is broadly motivated by the desire to understand how environmental factors shape organismal evolution and phenotypic expression. Our questions span scales of inquiry from genes to communities, and we examine evolutionary and ecological processes in both natural and agricultural systems. We use a diverse set of tools, including manipulative experiments, mathematical modeling, and genomics/transcriptomics. Current and future lab directions are listed below:
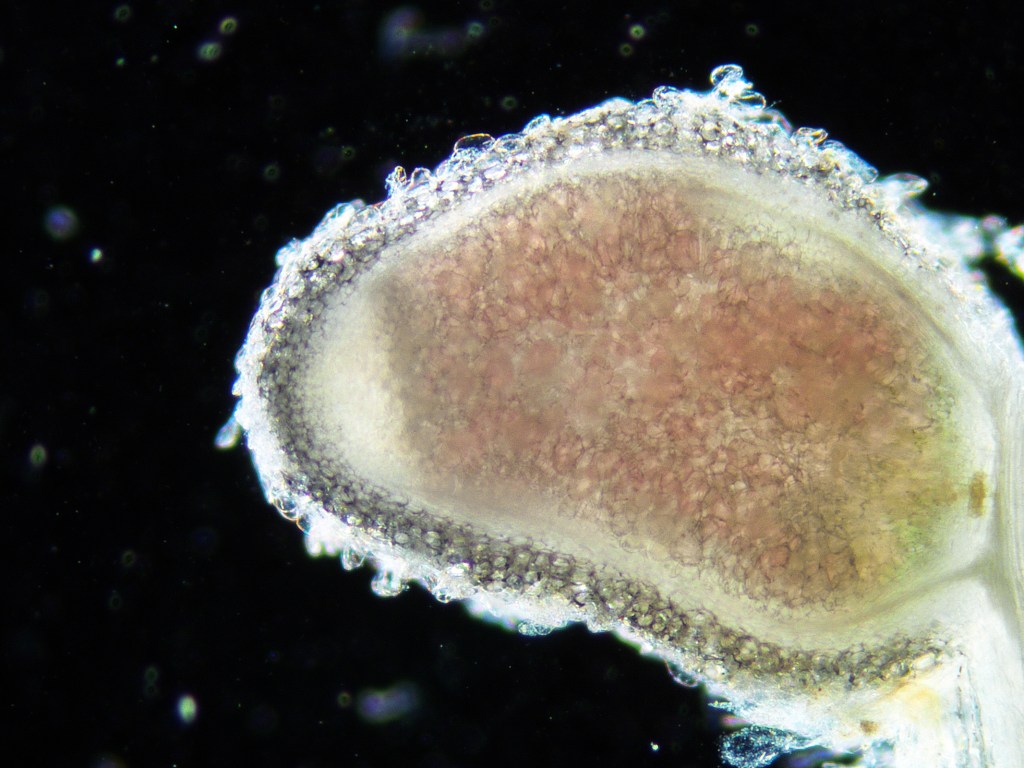
Evolutionary ecology of the mutualism between legumes and rhizobia
The evolutionary processes that maintain mutualism and the ecological processes that maintain genetic variation within mutualistic species have long fascinated scientists. By measuring the effects of environmental variation on the fitness of both partners across generations, we are moving closer to understanding the ‘rules of life’ governing mutualism.
Key papers: Burghardt 2019 (New Phytologist), Burghardt et al 2019 (Evolution), Batstone 2022 (Proc B)

Application of Select and Resequence approach for assessing strain fitness
In close collaboration with Peter Tiffin and Brendan Epstein, we’ve developed a method to use mixed inoculations of scores of strains and high-coverage genome sequencing to measure selection on rhizobia in host nodules and in the soil. Genome-wide association analysis can be used to identify natural genetic variation in rhizobia genes putatively underlying the measured fitness differences. This project is funded by NSF and DOE.
Key papers: Burghardt et al 2018 (PNAS), Epstein et al. 2018 (mSphere), Burghardt et. al 2022 ( AEM)

Functional genomics of host x strain interactions
The function of host and rhizobial symbiotic genes are often tested using only one or, at most, a few genotypes of the other species. We are working to change that by using S&R screens on host mutants and testing bacterial mutants in host competition experiments. This project is funded by a PGRP from NSF. We are also digging into measuring strain-induced changes in nodule and root architecture.
Key paper: Burghardt, Trujillo, 2019 et al. (Plant Physiology ), and Epstein et al. 2022 (Molecular Ecology)

Agricultural applications: Beneficial inoculants and leguminous cover crops
With new USDA funding and a new rhizobial collection specific to PA, the lab is expanding into thinking about the applied aspects of our work:
- Evolution-enabled management of beneficial inoculants
- Symbiont-informed legume crop development
- Use of cover crops for soil health and for maintenance of specific rhizobial strains
- Perennial intercropping

Plant and rhizobial life-cycle evolution
Phenological transitions like flowering and germination determine what environments plants are exposed to and thus selection. They also determine when rhizobia are inside host nodules and when they are in the soil. In her dissertation, L. Burghardt used tools from crop modeling to predict the life-cyles of different A. thaliana genotypes over time and space. Moving forward, we will use individual-based modeling to explore the interplay of plant and rhizobial life cycles.
Key papers: Burghardt et al. 2015 American Naturalist; Burghardt 2019 (New Phytologist)

Lab Outreach and Extension
We enjoy discussing our work with the public, farmers, and policymakers. Legumes, rhizobia and the climate measures that shape them are important, and we try to communicate this importance to a wide audience, to spark curiosity and to inform decision making. While I haven’t set up shop in State College, see the outreach page for info about of the prior outreach Burghardt has been involved in with the Tiffin Lab.
